Given an object of class "tracklets" containing a list of tracklets and including the value of turning angles, distance traveled and activity states (either 1 or 0 for active or inactive state respectively), this function compute the net square displacement value used to infer populations spread according to the equations derived from Turchin (2015) or Kareiva & Shigesada (1983). In a nutshell the Turchin's equation is a simplified version of the Kareiva & Shigesada' equation, assuming symmetric turning angles.
Usage
DCoef(
trackDat,
turnAngle = NULL,
distTraveled = NULL,
activityStates = NULL,
method = c("Turchin", "KS")
)Arguments
- trackDat
An object of class "tracklets" containing a list of tracklets and their characteristics classically used for further computations (at least x.pos, y.pos, frame).
- turnAngle
A character string indicating the name of the variable specifying the turning angles over each trajectories.
- distTraveled
A character string indicating the name of the variable specifying the distance traveled by the particles.
- activityStates
A character string indicating the name of the variable specifying activity state (coded as 1 or 0 for active and inactive state, respectively).
- method
A character string indicating the method used to compute the net square displacement, either "Turchin" or "KS" to use the computation derived from the equations of Turchin's book 2015 edition (corrected version of the chapter 5.3 from 1998) or from Kareiva & Shigesada (1983) (default = "Turchin").
Value
this function returns the net square displacement value used to infer populations spread according to the equations derived from Turchin (2015) or Kareiva & Shigesada (1983).
References
Kareiva, P.M., Shigesada, N., (1983). Analyzing insect movement as correlated random walk. Oecologia 56,234–238. https://doi.org/10.1007/BF00379695
Turchin, P., (2015). Quantitative Analysis of Movement: Measuring and Modeling Population Redistribution in Animals and Plants. Beresta Books, Storrs, Connecticut.
Examples
# simulate a correlated random walk with known parameter to verify Turchin D computation
## specify some parameters
nn = 1000
stepLength = 1
angularErrorSd = pi / 4
## create a function to simulate correlated random walk
myccrw <- function() {
disps = runif(nn, min = 0, max = 2 * stepLength)
angulos = runif(nn, min = -angularErrorSd, max = angularErrorSd)
lesx = rep(0, nn)
lesy = rep(0, nn)
curangle = runif(1, min = -pi, max = pi)
for (t in 2:nn) {
curangle = curangle + angulos[t]
lesx[t] = lesx[t - 1] + sin(curangle)
lesy[t] = lesy[t - 1] + cos(curangle)
}
return(data.frame(
x.pos = lesx + 250,
y.pos = lesy + 250,
frame = 1:nn
))
}
# simulated 30 tracklets
nbr = 30
simus <- MoveR::trackletsClass(lapply(1:nbr, function(rrr)
myccrw()))
# take a look at the simulated data
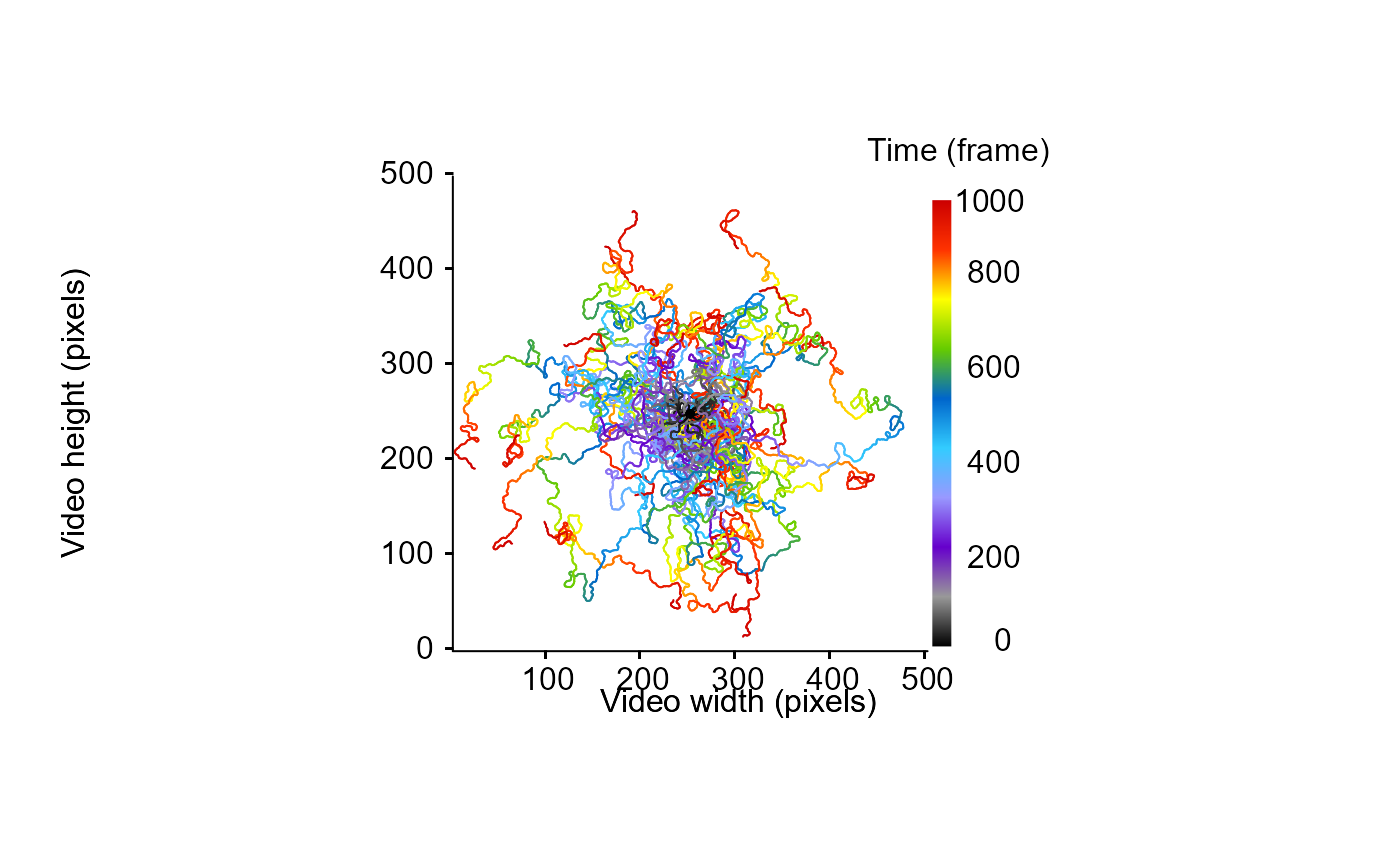
MoveR::drawTracklets(simus)
 # compute the needed metrics on simulated tracklets
simComp <-
MoveR::analyseTracklets(
simus,
customFunc = list(
## compute turning angle in radians over each tracklet (a modulus present within the MoveR package)
turnAngle = function(x)
MoveR::turnAngle(
x,
timeCol = "frame",
unit = "radians",
scale = 1
),
## compute distance traveled
distTraveled = function(x)
MoveR::distTraveled(x, step = 1),
## add behavioral states (consider as active all the time)
activity = function(x)
rep(1, nrow(x))
)
)
# compute the coeficient D corresponding to the value of net square displacement of the population
D <- MoveR::DCoef(
simComp,
turnAngle = "turnAngle",
distTraveled = "distTraveled",
activityStates = "activity",
method = "Turchin"
)
# compute the needed metrics on simulated tracklets
simComp <-
MoveR::analyseTracklets(
simus,
customFunc = list(
## compute turning angle in radians over each tracklet (a modulus present within the MoveR package)
turnAngle = function(x)
MoveR::turnAngle(
x,
timeCol = "frame",
unit = "radians",
scale = 1
),
## compute distance traveled
distTraveled = function(x)
MoveR::distTraveled(x, step = 1),
## add behavioral states (consider as active all the time)
activity = function(x)
rep(1, nrow(x))
)
)
# compute the coeficient D corresponding to the value of net square displacement of the population
D <- MoveR::DCoef(
simComp,
turnAngle = "turnAngle",
distTraveled = "distTraveled",
activityStates = "activity",
method = "Turchin"
)